|
The Strophariaceae ("Pholiostrohyphopsilocybe") [ Basidiomycetes > Agaricales . . . ] by Michael Kuo It has long been suspected that the mushrooms in Pholiota, Stropharia, Hypholoma, and Psilocybe share a common ancestor. However, since you and I also share a common ancestor with these mushrooms (a very different one, of course), the important question has to do with how recently this "Pholiostrohyphopsilocybe" appeared in the woods. It probably goes without saying that "1836" is not the answer, but I mention this silly possibility in order to point out that what is crucial in answering the question is not providing an exact date, but determining when Pholiostrohyphopsilocybe existed relative to the common ancestors of other mushrooms. If, for example, we were to discover that species of Amanita shared an ancestor with Pholiota, Stropharia, Hypholoma, and Psilocybe--a more recent ancestor than Pholiostrohyphopsilocybe--we would know that Pholiostrohyphopsilocybe was not just a ridiculous mouthful, but an inaccurate label that did not reflect the evolution of the mushrooms (in which case "Amanipholiostrohyphopsilocybe" might work). Mycologists used to have a hard time answering questions like these. Anyone who has seen the blackened, splatty mass of a former mushroom in the woods might guess that there aren't a lot of mushroom fossils found by scientists. Think of the animal skeleton you may have seen on the forest floor a few feet away from the ex-mushroom, add a few million years, and you can see why zoologists have a better paleontological record than mycologists. Instead of consulting fossils, mycologists over the years have been forced to theorize the evolution of mushrooms on the basis of the physical features of today's mushrooms, using a technique we might call triangulation. For a brief example of this technique,¹ consider these words in English, Spanish, and French:
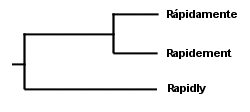
Rápidamente Rapidement I did not italicize the foreign words, as is the custom in English, because doing so would have coincidentally emphasized what is probably obvious without the italics: not only are the three words clearly related, but the Spanish and French words are clearly related more closely to each other than they are to the English word. However, step back and make sure you are thinking like a scientist; the Spanish and French words are not more closely related simply as a result of their foreignness (which is what the italics would have represented). Rather, they are more closely related because they differ on fewer characters than they differ from the English word. Simply by observing the features of the three words, then, we might theorize that they share a common ancestor, but that the Spanish and French words share a more recent common ancestor. A group of things that shares a common ancestor is called a "clade," and we have thus theorized two clades in the evolution of languages. We might make a "cladogram" that looks like something like this:
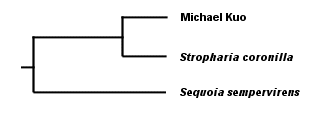
. . . in which our first clade (should we call it "Frenglanish?") contains all three words, and our second clade ("Frenish?" "Sprench?") is nested within the first clade and contains the Spanish and French words. Notice that we were able to theorize (accurately, in fact, as far as these words are concerned) about the evolution of languages without reference to fossils--even though at least one fossil was available to us in this case in the form of the Latin word rapidus. But using this method to theorize the evolution of mushrooms, based on observation of their physical features (with and without a microscope) has obvious problems. Do two mushrooms that have ornamented spores share a more recent common ancestor with each other than they do with a mushroom whose spores are not ornamented? Are two mushrooms with dark spore prints more closely related to each other than to a mushroom with a white spore print? You can see where this is heading. There are many characters to assess, and we have no sense of which characters are actually indicative of significant evolutionary change. Actually, the same problem existed with our deceptively simple example of Rapidly, Rápidamente, and Rapidement. What if the accent over the a in the Spanish word turned out to be a more significant difference than the difference between the suffixes -ly, -ment, and -mente? Or, what about the possibility that the Spanish and French words evolved the -ment in their suffixes independently? We have not proved that Spanish and French did not both evolve from English and develop superficial similarities merely by coincidence--or, for that matter, that the three words are necessarily related at all. You can see how seductive such arrangements as the one in our cladogram above can be, and why mycologists over the years may have clung stubbornly, for example, to the idea that two black-spored mushrooms whose gills liquify at maturity, Coprinus comatus and Coprinus atramentarius, share a more recent common ancestor than they do with the white-spored, sturdy-gilled Macrolepiota procera. The relationships between Coprinus and Macrolepiota seem obvious, and they just "make sense." Except that they're inaccurate. Mycologists now have a far more reliable means of deducing the evolution of mushrooms, and DNA studies of mushrooms are revealing some relationships that were previously unsuspected. I think it is important to note, however, that the theoretical method used by DNA studies is not all that different from the kind of triangulation we have been examining so far; it is just that the data being triangulated is more reliable. While we were not sure, above, which characters in our data set were actually informative (the accent mark in Spanish, the spore print color, and so on), we can be very sure that differences and similarities in the genetic code of mushrooms are indicative of their evolution. The DNA molecule of any organism is very large, and it contains, in code, the entire history of the organism's evolution. The challenge, obviously, involves how to read the code. Perhaps my bias as an English teacher leads me to say this, but I think that the interpretation of DNA data is the real issue, and that understanding the technology involved is relatively unimportant. We tend to get bogged down in our thinking, when considering DNA data, because the technology and methods generating the data seem so impenetrable. But who cares? They use some enzymes, some dyes, a machine with a laser, and a computer. Unless you're someone who can't drive to the store without being able to build a combustion engine in your garage, let's move on to what matters, which is how molecular biologists intepret the coded story in the DNA molecule. Since the DNA molecule contains the complete evolutionary story of an organism, it follows that some portion of the code will be the same for any two organisms. If we read my DNA and the DNA of Stropharia coronilla, a portion of the code will overlap. In discovering this, we have discovered a clade ("Kuopharia?"), and we have supported the idea that Stropharia coronilla and I share a common ancestor, represented by the shared portions of our DNA. If we now test the DNA of the redwood tree, Sequoia sempervirens, and find that it shares less genetic code than Stropharia coronilla and I do, we know that I am more closely related to the mushroom than the tree (this is in fact the case), and that the Kuopharian ancestor took another evolutionary road after having diverged from the common ancestor of the redwood, the Stropharia, and me:
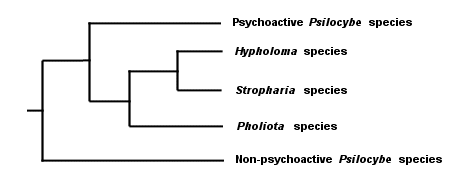
Do you see why I said that the triangulation method hasn't really changed? The difference is, we're now triangulating the DNA of these organisms, rather than their spore prints, colors, leaves, bones, cystidia, and so on. And, though I have said this elsewhere, it bears repeating: notice that we are able to deduce evolution reliably without fossils, and be sure to mention this the next time some idiot wants to teach your children "intelligent design" theory because evolution involves "gaps in the fossil record" and therefore can't be trusted. Notice, too, that reading the code of DNA does not necessarily tell us anything about taxonomy. The DNA results tell us simply that Stropharia and I share a more recent common ancestor than we do with the redwood. What we choose to call the common ancestors has nothing to do with the results. Neither does the question of whether Stropharia coronilla and I are different species, or whether we belong in the same genus, family, order, and so on--since the very idea of a "species" is precisely that, an idea, rather than a natural category defined by DNA. We can throw in results from an ascomycete like Peziza repanda, and we will discover that it shares an even more recent common ancestor with the Stropharia--and if we add results for Hypholoma sublateritium, yet another more recent ancestor to Stropharia coronilla will be discovered. For that matter, if we pick two specimens of Stropharia coronilla and test them, we will find that there are regions of difference in their DNA--just as you and I, though we are both human, have a portion of DNA that differs. How to line up DNA results with labels for individuals, species, genera, and so on is a question entirely unrelated to the results themselves. This is why some molecular biologists have proposed abandoning the project altogether and simply referring, instead, to clades. It is also why no one really knows what to call the various mushrooms that used to be called species of Pholiota, Stropharia, Hypholoma, and Psilocybe. DNA testing by Moncalvo and others (2002; citation and link below) of some of these mushrooms has revealed that our previous guesswork, based on triangulating their physical features, wasn't completely off-base, and that the cladogram for the mushrooms, greatly oversimplified, looks something like this:
I have left out a lot of details (including some results for a few species that did not line up where they "should have," based on our previous assumptions) so that we can focus on the big picture; follow the Moncalvo et al. link below for the complete cladogram (see page 368 of the PDF file). Also: the length of the lines in my cladograms is not meant to portray anything significant, though line length does have meaning in "real" cladograms; molecular biologists are able to make very good guesses about when genetic events occurred by using a "molecular clock"--but that is a topic for another day. Though we are not always this lucky, in this case interpreting the coded DNA of the mushrooms has confirmed many of our previous assumptions about relationships between them. In fact the only "big" error, based on the mushrooms that were studied, appears to have been our concept of the genus Psilocybe--which, it turns out, was too broad and should not have included the psychoactive species (or at least the ones that were studied by Moncalvo and his coauthors). Hypholoma and Stropharia are the most closely related in the cladogram, sharing the most recent common ancestor. In turn these genera are more closely related to Pholiota than they are to the psychoactive species of Psilocybe, while the non-psychoactive species of Psilocybe diverged from the path long before any of the other divergences. But are Hypholoma and Stropharia really separate genera? Should we call Hypholoma, Stropharia, and Pholiota a family? The DNA results do not answer these questions, and they are not going to provide answers no matter how many mushrooms we test--because these are questions about language and labels, not DNA. To be honest, I don't particularly care what the mushrooms are called, or at what level we decide the differences in genetic code reflect differences in individuals, species, genera, and so on. What I find fascinating instead, for example, is the fact that most of the Stropharia species are saprobes on grass, dung, or forest litter, while the mushrooms in Pholiota and Hypholoma are saprobes on decaying wood. This tells me that the common ancestor of Stropharia, Hypholoma, and Pholiota was likely a wood-rotter, and that Stropharia branched away in an adaptation to a different ecological role. Consider how important this idea might be. If we can figure out when, where, and why Stropharia split away from its wood-rotting cousins in order to decompose grass, dung, and forest litter, we might discover something truly significant, not only for mycology, but for geology, plant biology, and zoology as well. Here is one of several hypotheses that comes to mind: the common ancestor was a forest wood-rotter, but part of the population was separated by a geological event that left the future Stropharia clade isolated. Something cataclysmic--say, a tsunami--happened in the isolated area, killing all the trees but leaving the grasses to flourish in the sandy soil; Stropharia adapted. Eventually, Stropharia (or part of it) traveled again, and was reintroduced to the forest lifestyle, where its new trade (decomposing litter) suited it quite well on the forest floor and did not require another adaptation to re-acquire wood-rotting ability. Yes, I made up that scenario without even thinking about it very much, and it probably wouldn't hold up under much scrutiny. But the point is that this kind of theorizing and thinking is much more interesting and important than squabbling over whether Stropharia coronilla is really Psilocybe coronilla. This is why DNA studies of mushrooms are so exciting! Let's not miss out on seeing the forest by focusing too much on the But we have to call them something! Yes, we do. So let's use the names we've been using, amending them as necessary in order to reflect all the fascinating new things we are finding out. We will of course have to shift names around, create new ones, and so on. But there's no point in getting all worked up about the names we choose. Moncalvo and coauthors have applied the name "Stropharioid Clade" to the top four branches of the cladogram above, and have used the genus names Hypholoma, Stropharia, and Pholiota to represent the second, third, and fourth clades. The top clade, consisting of the psychoactive members of Psilocybe, they have labeled the "Psychadelia Clade," while the bottom clade (the rest of what used to be Psilocybe) gets the name Psilocybe. If you want to quibble, feel free. Meanwhile, I will use the genus names Hypholoma, Stropharia, and Pholiota--and worry about what to do with "Psilocybe" when I or one of this Web site's contributing authors find one and make a page for it. At the family level, I have elected to put Hypholoma, Stropharia, and Pholiota in the "Strophariaceae," corresponding to what Moncalvo et al. call the "Stropharioid Clade." The current Dictionary of the Fungi, incidentally, puts all of these mushrooms in the Strophariaceae, and treats all the genus names as synonyms for Psilocybe--but the edition was published prior to the studies we have been looking at. ¹ I am indebted to Richard Dawkins (The Ancestor's Tale, 2004) for the idea of applying a cladogram to linguistic, rather than genetic, evolution. References Kirk, P.M. et al., eds. (2001). Ainsworth & Bisby's dictionary of the fungi. Oxford: CAB International. 655 pp. Moncalvo, J. M., et al. (2002). One hundred and seventeen clades of euagarics. Molecular Phylogenetics and Evolution 23: 357–400. An online version of this paper is available at: http://www.biology.duke.edu/fungi/mycolab/publications/117clades.html Cite this page as: Kuo, M. (2005, January). The Strophariaceae ("Pholiostrohyphopsilocybe"). Retrieved from the MushroomExpert.Com Web site: http://www.mushroomexpert.com/strophariaceae.html © MushroomExpert.Com |