| Major Groups > Stinkhorns > Blumenavia angolensis |

|
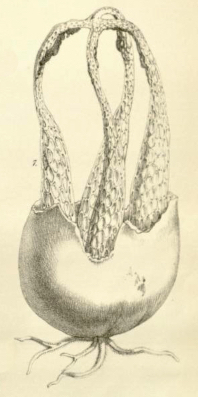
Blumenavia angolensis [ Agaricomycetes > Phallales > Clathraceae > Blumenavia . . . ] by Michael Kuo The name "Blumenavia angolensis" is used frequently in field guides and in stinkhorn literature, but it is probably misapplied in many of these sources. Often a better name would be Blumenavia rhacodes, but see the linked page for other possibilities. As the species epithet suggests, Blumenavia angolensis was originally described from Angola; that description (Welwitsch & Currey 1868) appears below, and the illustration from the description appears to the right. While the description is brief, it is clear about the bright white color of the stinkhorn's arms, and its dark volva. I have had photos sent to me from the Gauteng Province of South Africa (roughly 1500 miles from Catete, Angola, where the original collection was made) that appear to match this description—but the photographed specimen was not preserved, so it cannot be studied and, in the interests of not propagating an error I am about to delineate (see the review below), I will refrain from reproducing the photos here. Blumenavia angolensis has not been adequately studied in a contemporary, DNA-informed setting, and one recent publication in particular has unfortunately made a real hash of things; see the review below if you care. Have you collected something in Africa that looks like Blumenavia angolensis recently? I would love to study well-documented, preserved collections, in order to make this page—and the contemporary concept of the species—more scientific. If you're interested in helping, please send me an email at . Description: Note: This description is from the protologue (Welwitsch & Currey 1868). The original publication is in Latin and English; I have translated the Latin. LATERNEA ANGOLENSIS, Welw. & Curr. Peridium gray, blackish spotted, very thick, filled with gelatin; receptacle four-armed, bright white, joined at the apex; mucous layer gray-black, odor of fermented grapes. Hab. On sandy rocks covered with soil at the Garrison, Catete, Pungo Andongo, 10 Dec. 1856. No. 120. Tab 17, fig. 7, the plant, natural size, after a sketch taken on the spot. Obs. A single specimen, gathered in fine weather. This species is manifestly allied to Laternea columnata, Bose, but differs in the colour, and apparently also in the form of the ramifications of the receptacle, as well as in the smell (not fetid) of the mucous layer. |
|
|
Review of "Diversity in Trapped Cages" (Melanda et al. 2020)
This paper provides tentative support for recognition of several species in the genus Blumenavia, but is, ultimately, highly problematic. Because the problems with the research are illustrative of several points that I find myself making regularly in other contexts, I am publishing this review here in order to delineate and explore these points—not as a rejoinder to the authors themselves (I know none of them, but my library contains works by many of them, including some very good research). Point 1: No voucher? Oucher. The principle here is one that almost everyone learned in grade-school science class: if the experiment cannot be repeated, it isn't science. The authors of "Diversity in Trapped Cages" have used a DNA sequence, downloaded from GenBank (the primary international depository for DNA sequences), representing "Blumenavia angolensis." The sequence, representing the LSU ("Large Subunit") gene, was deposited in GenBank by Belgian mycologists (Degreef and collaborators) in 2012 as they prepared a manuscript that was published in 2013 ("Two rare Phallales recorded from São Tomé"). The sequence was obtained from collection "JD772" (presumably made by Jérôme Degreef, the first author), according to the publications and the GenBank record. You can examine this record yourself by going to GenBank and entering the GenBank sequence number—KC128653—in the search box. Under "FEATURES: source," you can find the data entered by the authors about the collection; the DNA sequence itself is below that, under "ORIGIN." Notice, also, that Degreef and collaborators put the collection number (JD772) in the overall title for the sequence, which appears in bold letters at the top of the page: "Blumenavia angolensis isolate JD772 28S ribosomal RNA gene, partial sequence." Including the collection number in the title, while not required by GenBank, is something many scientists do to make searching the database easier. In the Materials and Methods section of their paper, Degreef and collaborators say that "[t]he collections are deposited at the National Botanic Garden of Belgium's herbarium," which means that JD772 is available for other researchers, making the experiment (extraction of DNA and sequencing the LSU gene) repeatable. So far, so good. But something apparently happened to the collection between 2012 and 2020, because Melanda and collaborators report that they "contacted Dr. Jerome [SIC] Degreef (Belgium) to study the specimen described in Degreef et al., since there is an available LSU sequence in GenBank; Dr. Degreef confirmed to us that the specimen was not located, nor the DNA isolate, thus it was not possible to obtain sequences from more loci." In other words, JD772 has gone missing. Perhaps someone at the National Botanic Garden of Belgium filed it away in the wrong cabinet. Perhaps some mycologist borrowed it and never returned it, and the paperwork got lost. Who knows. But the experiment is now not repeatable, so use of the sequence is not science. Collections get lost; there's really no avoiding this reality. I have never been to the National Botanic Garden of Belgium's herbarium, but I have visited plenty of large herbaria, some of which look a lot like the huge room at the end of Raiders of the Lost Ark, in which the Ark of the Covenant is filed away. Records and specimens in such places are easily confused, misplaced, and so on. Because GenBank sequences are publicly available, mycologists use them in their DNA alignments—and once a sequence appears in someone's paper, future papers tend to repeat use of the sequence. But what if the collection was misidentified? Without a voucher (a collection placed in a herbarium) we have no way of supporting (or contradicting) the DNA sequence with further research. What if the herbarium burns to the ground? This scenario has actually unfolded and created a huge mess with bolete research: sequences from decades ago appear in GenBank and are used and re-used in bolete studies, over and over, but the vouchers no longer exist because they were placed in a German herbarium that was later destroyed by fire. Rolls eyes emoji. Melanda and collaborators made a commendable, good faith effort to repeat the experiment and study JD772 themselves—and when the voucher was unavailable, they duly included this information in their paper. But here's the rub: data from the sequence should not have been used. Instead, the researchers used the sequence in their alignments, and drew taxonomic conclusions based on the position of the sequence in their phylogram (the DNA "tree"). I'll repeat: What if the mushroom was misidentified? And: What happens when the software does the alignment without the sequence? Do the same branches get produced? Point 2: "Multigene" phylogenies require . . . well, multiple genes Take a look at Table 1 from Melanda and collaborators. The five columns on the right represent genes; the authors sequenced five different genes from collections included in the paper, then asked the software to sort everything out and calculate relationships between the mushrooms. But in the top row of the table we discover that "Blumenavia angolensis" is represented by only one gene sequence, the LSU gene, obtained from our old friend JD772, deposited in GenBank as KC128653. Inputting this sequence into the software is completely useless when the other species are represented by four and five genes. You can verify this point for yourself, and have a BLAST while doing so: Return to GenBank and follow the link to "BLAST" (Basic Local Alignment Search Tool); then select "Nucleotide BLAST." In the "Enter accession numbers" box, type in KC128653 (the GenBank number for JD772), then click BLAST. It may take a while for your results, but you should eventually see a list of "Sequences producing significant alignments." The column "Per. Ident" shows you what sequences in the GenBank database match the identity of JD772, by what percent. Since there may be additional relevant sequences in the database by the time you read this, I'll not use precise locators like "the first row," but you should see several LSU sequences that share 100.00 percent identity with JD772. In other words, all 616 base pairs of As, Cs, Gs, and Ts occur in exactly the same order. The sequence is identical to LSU sequences, found in other rows, for mushrooms identified as Blumenavia baturitensis and Blumenavia rhacodes! A quick aside for some background information: The LSU gene was one of the first genes used by mycologists in DNA studies, especially in the 1990s and early 2000s, when many papers were published based on LSU phylogenies. But further research and experience led mycology to the discovery that LSU is more useful for painting with broad strokes than for finely detailed work. Thus LSU is more informative when it comes to identifying, say, a mushroom's genus or family—and less informative for separating species. If LSU indicates that two mushrooms represent different species, the likelihood is high, since the broad-strokes painting shows the separation. But if LSU indicates that the two mushrooms are the same, we can only have limited confidence in the result, as we saw above, and we should consult more genes. This is one of many reasons that "multigene phylogenies" are the standard at the time of this writing. Back to the "Diversity in Trapped Cages" paper: now you can see why including a species represented by only one gene is ridiculous. In Figure 1 we find "B. angolensis," represented by a single LSU sequence of JD772, on its own line. But the computer had no idea what to do with the sequence (as is evidenced by the total absence of support for the node that breaks the sequence onto its own branch, "-/-/-") and the mushroom could have been placed virtually anywhere in the tree, since that sequence is, as we discovered, 100 percent identical to LSU sequences for two of the other species in the phylogram. Never mind for the moment the fact that JD772 does not exist; even if it did exist the LSU sequence should not have been used alone. Presumably, the authors intended to sequence the other 4 genes from JD772, which is why they contacted Degreef. But scientific intentions are not a substitute for science itself, and inclusion of the sequence is a misrepresentation of what we actually know about Blumenavia angolensis based on the authors' research, which is this: a sequence labeled "Blumenavia angolensis," downloaded from GenBank, representing a missing mushroom that no one can ever study again and a single gene that is not particularly informative for species determination, is 100 percent the same as sequences for at least two other Blumenavia species. Conclusion To the authors' credit, these problems (use of a sequence that was unvouchered, and including a single LSU sequence to represent a taxon in a multigene phylogeny) were easily discovered in the paper itself, without much digging—which perhaps means that the paper's peer reviewers bear some of the responsibility for the errors. This particular journal makes its peer review commentary publicly available (click the "Peer Review" tab on the article); I will be generous and say the peer review was, um, minimal. What should have been done, instead? Blumenavia angolensis should have been treated as an uncertain entity rather than represented as a species supported by the paper's research. The sequence should not have appeared in the phylogram. Since the paper purports to be a "revision" of Blumenavia, discussion of Blumenavia angolensis should have been included under a label like "Uncertain Taxa." Acknowledgments Thank you to the professional mycologists who helped me prepare this review. REFERENCES: (F. M. J. Welwitsch & F. Currey, 1870) D. M. Dring, 1980. (Möller, 1895; Lloyd, 1905; Dring, 1980; Vargas-Rodriquez & Vázquez-García, 2005; Degreef et al., 2013; Rodrigues & Baseia, 2013; Trierveiler-Pereira et al., 2014; Desjardin & Perry, 2015; Melanda et al., 2020.) This site contains no information about the edibility or toxicity of mushrooms. Cite this page as: Kuo, M. (2022, May). Blumenavia angolensis. Retrieved from the MushroomExpert.Com Web site: http://www.mushroomexpert.com/blumenavia_angolensis.html © MushroomExpert.Com |